Advertisement
If you have a new account but are having problems posting or verifying your account, please email us on hello@boards.ie for help. Thanks :)
Hello all! Please ensure that you are posting a new thread or question in the appropriate forum. The Feedback forum is overwhelmed with questions that are having to be moved elsewhere. If you need help to verify your account contact hello@boards.ie
Yamna culture DNA
Options
-
18-01-2012 4:42pm#1I was just wondering if anybody on this forum has any knowledge of the DNA of the Yamna culture, specifically the extraction of ancient DNA from burial mounds. In a broader context any of the Steppe Kurgan cultures would be interesting to know about.
Since the Yamna culture and/or related Steppe cultures are almost certainly the people who spoke Proto-Indo-European I was wondering what is known about them genetically.0
Comments
-
I was just wondering if anybody on this forum has any knowledge of the DNA of the Yamna culture, specifically the extraction of ancient DNA from burial mounds. In a broader context any of the Steppe Kurgan cultures would be interesting to know about.
Since the Yamna culture and/or related Steppe cultures are almost certainly the people who spoke Proto-Indo-European I was wondering what is known about them genetically.
Of the top of my head I haven't kept up a huge amount regarding ancient-DNA and the steppes. Most of the current data is pointing towards Haplogroup R1a as connected. I know the Johannes Gutenberg University Mainz (Germany) have a project that is examing ancient DNA (aDNA) from the region. See:
http://www.uni-mainz.de/FB/Biologie/Anthropologie/MolA/English/Research/CentralAsia.html
One of the Kurgan's excavated did have viable aDNA which gave a result of Y-Haplogroup R1a1. I believe it's date to 1800-1400BC see list of ancient DNA results here:
http://www.buildinghistory.org/distantpast/ancientdna.shtml
Haplogroup R* probably fist arose in Central Asia about 25k years ago. It's child haplogroups R1a and R1b have high correalation with Indo-European speaking populations.
Tarim basin mummies which are linked to the Tocharians (IE branch) have had a number come back as R1a1a (Males)0 -
I should add here is a map proposing locations of where different SNP's arose. Different clades (R1a vs. R1b) are marked by different SNP's.
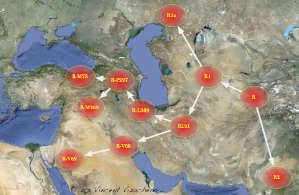
R1b-M269 is the dominant clade in western Europe, nearly all western European R1b belongs to it's subclades. (close on 80-90% of Irishmen for example)0 -
I should add here's a very usefull Phylogenetic tree of Haplogroup R1a based off current know distrubiton of it's subclades. I got it from the R1a discussion group on DNA-Forums.

Obviously not fully on topic but it does show that R1a for example has high incidence in groups that speak Satem languages. Namely:- Indic
- Iranic
- Salvic
- Baltic
The spread into Germanic populations appears to be due to mix of an early migration into Scandinavia (which is one reason R1a in Ireland is connected to Vikings) and that Germany forms boundary area between highest areas of R1a and R1b frequency. As a result Germans have high level of both R1a and R1b.
Italic/Celtic speaking populations in general have high levels of R1b. Obviously they and Germanic are Centum languages.0 -
Extremely interesting dubhthach!
Of particular interest is the genetic reflection of linguistic relations. The fact that the Centum and Satem language groups grouping together in some way.
Of course there is some confusion here from a linguistic perspective in that we aren't sure that Centum and Satem reflect anything about Proto-Indo-European's development. Greek and Indo-Iranian both evolved from Late Indo-European, but one is Centum and the other Satem. It seems each branch became Centum or Satem independently*. Although we can say that late Indo-European languages had a tendency to become Satem.
However very interesting to know that Kurgan cultures seem to have R1a.
Would you have any knowledge of when R1a and R1b arose?
For anybody reading Centum and Satem refer to the specific way what are known as "the velar consonants" evolved in different Indo-European languages. The velar consonants are ones made with the back of your mouth and the back of your tongue, e.g. k in kick, g in goat, q in queen.
Indo-European had nine of these:
k k^ kw
g g^ gw
gh gh^ ghw
An example is k, k^ and q.
k - Typical English sound, the c in "Cat"
k^ - doesn't really exist in English, it's basically k made with the middle of your tongue touching the middle of the roof of your mouth.
kw - Is just q.
g - just g
g^ - not in English, g made with the middle of your mouth
gw - not in English, the easiest way to describe it is try to say gueen instead of queen and remember where your tongue was for queen. Or you'll notice your voice box shuts off when you go to the q in queen, keep it on instead.
The gh, e.t.c. are the same as g, you just release a burst of air after you say them. Similar to the burst of air after you say the P in Principle.
Basically Centum languages deleted the sounds with ^.
The Santem deleted the sounds with w and then turned the ^ sounds into s, sh or j sounds. s, sh and j are made with the middle of the mouth so they're "close" to these sounds but are much easier to make.
*Reference: Fortson, B.W. IV "Indo-European Language and Culture, An Introduction", 2nd Ed. p. 58-590 -
Would you have any knowledge of when R1a and R1b arose?
Well I've heard dates of about 18,000 years ago for the last common ancestor between modern R1a and R1b men. Obviously this is way too early for Indo-European. As a result there are R1b groups that are completely non-IE speaking. For example on the map above you can see R-V88/R-V69 (R1b1c* / R1b1c4 -- R1b-V69 is subclade of R1b-v88). It heavily concentrated in Africa among Chadic speakers. With a distribution from western middle east all the way over to Ghana.
Here's a direct copy from ISOGG's latest version of Haplogroup R tree:Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.- R1a-M420 is believed to have arisen on the Eurasian Steppe or the Indus Valley, and today is most frequently observed in eastern Europe and in western and central Asia. Haplogroup R1a1a1g-M458 is found at frequencies approaching or exceeding 30% in Eastern Europe.
- R1b-M343 is believed to have arisen in southwest Asia and today its sublcades are bound in various distributions across Eurasia and Africa.
- Paragroup R1b1* and haplogroup R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.
- Haplogroup R1b1a-P297 is found throughout Eurasia.
- Haplogroup R1b1a1-M73 is observed most frequently in Asia, with low frequency of observation in Europe.
- Haplogroup R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia. R1b1a2-M269 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup R1b1a2a1a1-L11/S127 and most European R1b1a2 belongs to haplogroups R1b1a2a1a1a-S21/U106 or R1b1a2a1a1b-P312/S116.
- Haplogroup R1b1a-P297 is found throughout Eurasia.
- Paragroup R1b1* and haplogroup R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.
Source: http://www.isogg.org/tree/ISOGG_HapgrpR.html
Now the Y-Chromosome only makes up about 2% of a man's genome. There have been studies looking at the whole genome and doing comparison's across all populations. These look for admixture between different ancestral components. One thing that has been noticed is that all Indo-European speaking populations appear to have an "ancestral component" that peaks in populations in the Caucasus. Interesting enough Basques have next to none of this "ancestral component" (though they have plenty of R1b from introgression of males).
As a result one theory that has been proposed is that is that Proto-IE arose on the pontic steppe and that early P-IE speakers (who may have been R1a) absorbed both a Caucasian population as well as groups that were R1b. The age of R1b-M269 for example is calculated at up to 8k years old.
One speculative map on spread of R1b into Europe with dates.
Current R1a1a* (including all subclades) distrubition in Eurasia. 0
0 -
Advertisement
-
Fascinating!Now the Y-Chromosome only makes up about 2% of a man's genome. There have been studies looking at the whole genome and doing comparison's across all populations. These look for admixture between different ancestral components. One thing that has been noticed is that all Indo-European speaking populations appear to have an "ancestral component" that peaks in populations in the Caucasus. Interesting enough Basques have next to none of this "ancestral component" (though they have plenty of R1b from introgression of males).
From a linguistic point of view this has to be the case, it's the only location the reconstructed vocabulary is consistent with. Good thing the genetics matches this.As a result one theory that has been proposed is that is that Proto-IE arose on the pontic steppe
No way! We know from linguistic analysis that Indo-European had strong contact with Proto-Kartvelian, this would seem to match that in some way. We also know from linguistics that it is possible that, not the Indo-Europeans themselves, but the Indo-Iranians possible subjugated the Proto-Finno-Ugric peoples. Any interesting genetics from the Finnish?and that early P-IE speakers (who may have been R1a) absorbed both a Caucasian population as well as groups that were R1b. The age of R1b-M269 for example is calculated at up to 8k years old.
Thanks for the info!0 -
Join Date:Posts: 4957
Is that R1b map pointing to clusters which have been identified or just outlining the general trend? There seem to be seven arrows ending in Britain.
Did R1b spread eastward?0 -
slowburner wrote: »Is that R1b map pointing to clusters which have been identified or just outlining the general trend? There seem to be seven arrows ending in Britain.
Did R1b spread eastward?
Slowburner that map is attempting to show general migration of different groups under R1b. There are some R1b samples know from pakistan and into the Tarim basin among a Uigher sample 3% were R1b+ this could imply descent from Tocharians. The Uighers are a very interesting group as they are a near 50/50 European/Asian admixture. As a result they have a full range of features from more Asian looking to nearly european looking.
Here are some R1b specific maps, unfortunatley I don't have one that maps more then just Europe and the very near east.
R1b-M269 (R1b1a2) -- poor map only areas with over 13% stand out
R1b-L11 (R1b1a2a1a1) -- subclade of M269 (all L11+ men are M269+)
As comparison here's map of all R1b-M269+ men who tested negative for L11 (and thus all of it's subclades)
Here is a tree from late last year showing R1b-L11 main groups, new finds have occurred in time since it was created.
U106 (S21) on the left has a strong correlation with Germanic speaking Europe, here is map:
U106/S21
P312 on the right has a closer correspondence with Italic-Celtic populations. Two of the major clades are L21 and U152 (S28)
U152 (S28)
L21
Some major discoverings under last 12months with new SNP's been discoverd that further divide up clades. In general though from an ancient history point of view I think there will be major developments in extracting viable DNA from ancient remains. This will give alot better picture. What we have at the moment only reflects the current population distrubition. It's only with samples from ancient remains that we can get better picture.
So far on neolithic remains in Europe no Haplogroup R* has been found. The oldest R1b sample been from Liechenstein cave in Germany and dating to 1000BC.0 -
Join Date:Posts: 4957
Is the U152 (S28) map telling us that there is no Italian genetic influence on Ireland?
It's pretty striking.0 -
slowburner wrote: »Is the U152 (S28) map telling us that there is no Italian genetic influence on Ireland?
It's pretty striking.
Well U152 appears to be connected to Alpine/Northern Italy. Some have proposed that it's specificly tied to La Tène material culture. The study that map has found U152 occurrence from 1.1% to 6.3% of men tested in Ireland. Here is breakdown of Ireland test areas based on major R1b clades.
Here are the samples with more then 50 men in them. I ignore the smaller ones as there would be too much of a margin of error.
West Ireland -- 67 samples- L21 = 73.1%
- U106 = 4.5%
- U152 = 1.5%
- P312 (non L21/ non U152) = 7.5%
South Ireland -- 89 samples- L21 = 74.2%
- U106 = 3.4%
- U152 = 1.1%
- P312 (non L21/ non U152) = 7.9%
East Ireland -- 149 samples- L21 = 71.1%
- U106 = 6.7%
- U152 = 4%
- P312 (non L21/ non U152) = 7.4%
North Ireland -- 72 samples- L21 = 79.2%
- U106 = 4.2%
- U152 = 1.4%
- P312 (non L21/ non U152) = 4.2%
This is taken from the supplementary data of:
The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269
http://rspb.royalsocietypublishing.org/content/early/2011/08/18/rspb.2011.1044.full.pdf
http://rspb.royalsocietypublishing.org/content/suppl/2011/08/18/rspb.2011.1044.DC1.html%20
Published in the "Proceedings of Royal Society -- Biological Sciences"
The report itself is arguring against Neolithic origin for R1b-M269 in Europe. However it's been argued that they are the "old-guard" that believe that R1b in Europe is a result of a mesolithic refuge in Spain (which is clearly wrong).0 -
Advertisement
-
not the Indo-Europeans themselves, but the Indo-Iranians possible subjugated the Proto-Finno-Ugric peoples. Any interesting genetics from the Finnish?
Thanks for the info!
Hey Enkidu, missed this part. Well Haplogroup N is generally associated with Finno-Ugric speakers. For example it reaches a frequency of 58% in Finland, though interesting enough it's only 1% of men in Hungary! (This could be tied to massive depopulation in Hungary during Mongol and later Ottoman invasions)
Another Siberian specific haplogroup is Q. Q is the brother of R* (paragroup containg R1a,R1b and R2), it's also the most common haplogroup among Native Americans.
If Finno-Uralic / siberian people had an affect on the "Indo-Iranians" then perhaps we would see elevated levels in them. On a quick search unfortantley I can only find data specific to Iran. None from Afghanistan, Tajikistan, Pakistan, India. (Need to do some digging).
Anyways on Iran sample I see 2.5% Haplogroup N and 4% Haplogroup Q. Now the Q sample could be due to influences coming in from Siberia/Central Asia in middle ages (Turks, Mongols etc.). As a comparison the Armenians have 0.5% Haplogroup N.0 -
There has been a couple of clusters created at the R1a1 + Subclades project for Irish British r1a members. My family fall into one of the clusters, if there is anybody here r1a1 with British, Irish ancestry joining the project would be very useful and an upgrade of SNP helps.0
-
Thanks guys - still confused about this, but now at a much deeper level0
Advertisement